MitoMicro Prin2020
MitoMicro – Mitochondrial MicroRNAs, a new perspective for mitochondrial function and their role in Eukaryotic evolution
Link to dedicated website: MitoMicro
(772.411 Eur, PI Marco Passamonti; Ilaria Negri; Francesco Nardi)
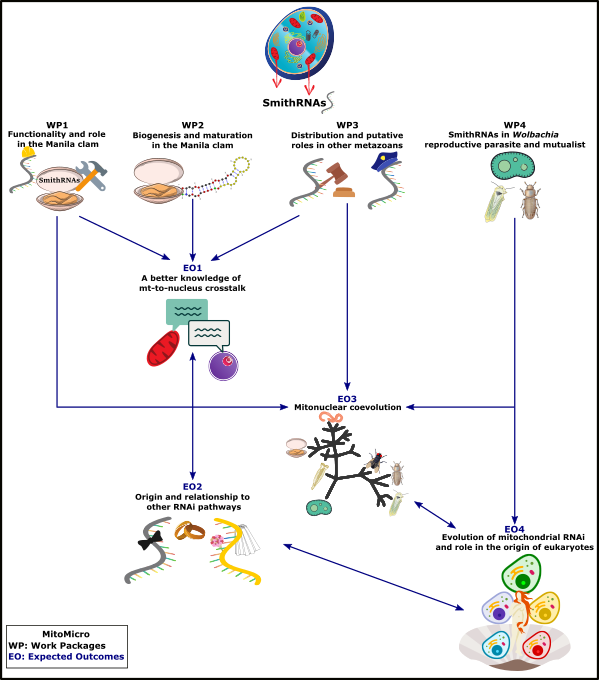
This project stems from our recent discovery of a new class of small non-coding RNAs (sncRNAs), called smithRNAs, coded by the mitochondrial genome (mtDNA) and predicted to regulate nuclear gene expression [1]. Functionality of at least two smithRNAs has been already proved in vivo [2], and more experiments are planned in this project. The possibility that mtDNA can act on nuclear genes regulation has never been suggested before, and it is likely to open a new avenue in mitochondrial studies. Therefore, the main objective of this project is to better characterize smithRNAs and their roles in eukaryotes. SmithRNAs have been first discovered in the Manila clam Ruditapes philippinarum where they are related to gonad formation and Doubly Uniparental Inheritance (DUI) [1]. We now know that they are found in some other animals as well, and that they could be involved in additional functions besides sex determination [2], but many aspects of their biology remain totally unstudied (biogenesis, target genes, taxonomic distribution, evolutionary conservation). Indeed, the smithRNAs may be regarded as a generalized new form of retrograde signaling (i.e., the mitochondria-to-nucleus signaling) and, potentially, a common feature among animals.
In detail, we want to boost this brand-new research field by:
1) an in-depth characterization of smithRNAs in R. philippinarum with selected functionality tests (WP1);
2) exploring their biogenesis, to assess if they pertain to siRNA, miRNA or piRNA, or rather to a new alternative class of small non-coding RNAs (WP2);
3) further assessing their presence in selected metazoan species and their evolutionary conservation (WP3); and
4) analyze the alpha-proteobacterium Wolbachia, known to be a sex distorter in arthropods, to assess whether a similar RNA interference mechanism is present in alpha-proteobacteria that are regarded as the closest “free-living” relatives of mitochondria (WP4).
If found, this will provide a far-reaching functional characterization of this new type of sncRNAs and an initial indication on their possible evolution, with a focus on endosymbiosis and the early stages of eukaryogenesis. Overall, this project will contribute to improve our knowledge on several aspects of mitochondrial biology and evolution, thus promoting a better understanding of mitochondrial function, interactions, gene regulation and evolution. The figure below show the project flowchart of MitoMicro.
Old Grants:
- PRIN2009: I sistemi a Doppia Eredità Uniparentale (DUI): un utile modello per lo studio dell’eredità e della genomica mitocondriale.
- PRIN2007: Struttura, evoluzione ed eredità del genoma mitocondriale dei Bivalvia.
- PRIN2005: Caratterizzazione di genomi mitocondriali in insetti pterigoti (Phasmatodea, Lepidoptera) e molluschi bivalvi.